Judith Reeks
Clustered regularly interspaced short palindromic repeats (CRISPRs) are a prokaryotic defence mechanism against horizontal gene transfer (HGT), which is in some ways comparable in function, but not in mechanism, to eukaryotic RNA interference. Insertion of foreign DNA and/or RNA from bacteriophage, viruses and conjugative plasmids into the cell results in cleavage of the non-native nucleic acid, thus preventing HGT. CRISPRs are arranged in an array of identical repeat sequences separated by short spacer sequences derived from previously encountered foreign DNA. The array acts as a "genetic memory" and provides immunity against the foreign genetic element from which the spacer was acquired.
Linked to the CRISPR array are CRISPR-associated (cas) genes, which encode for the proteins that mediate the CRISPR response to foreign nucleic acids. The response can be divided into three separate processes: (i) adaptation, the acquisition of a new spacer; (ii) expression, the transcription of the CRISPR array and subsequent processing of the RNA molecule to form mature CRISPR RNA (crRNA); (iii) interference, the destruction of foreign dsDNA or ssRNA. In collaboration with Malcolm White, we study the CRISPR systems of the hyperthermophilic Chrenoarchaeon Sulfolobus solfataricus. We use X-ray crystallography and other biophysical techniques to study the structure and function of Cas proteins and the roles they play in the complexes that mediate CRISPR function.
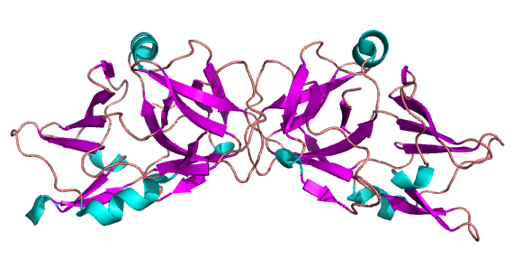
The prokaryotic clusters of regularly interspaced palindromic repeats (CRISPR) system utilizes genomically encoded CRISPR RNA (crRNA), derived from invading viruses and incorporated into ribonucleoprotein complexes with CRISPR-associated (CAS) proteins, to target and degrade viral DNA or RNA on subsequent infection. RNA is targeted by the CMR complex. In Sulfolobus solfataricus, this complex is composed of seven CAS protein subunits (Cmr1-7) and carries a diverse "payload" of targeting crRNA. The crystal structure of Cmr7 and low-resolution structure of the complex are presented. S. solfataricus CMR cleaves RNA targets in an endonucleolytic reaction at UA dinucleotides. This activity is dependent on the 8 nt repeat-derived 5' sequence in the crRNA, but not on the presence of a protospacer-associated motif (PAM) in the target. Both target and guide RNAs can be cleaved, although a single molecule of guide RNA can support the degradation of multiple targets.
Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, Naismith JH, Spagnolo L, White MF. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. (2012). Mol Cell. 45:303-13.

