Stephen McMahon, Lucile Moynie, Magnus Alphey, Huanting Liu
AEROPATH is an ambitious multidisciplinary research programme designed to support early stage antimicrobial drug development. We will advance understanding, at the molecular level, of fundamental and important aspects of the biology of Gram-negative bacteria targeting in particular the important pathogen Pseudomonas aeruginosa. The title is derived from P. aeruginosa pathogenesis.
Gram-negative bacteria present a particular problem in Europe. An ageing population, improved treatments for long-term human diseases such as diabetes and cancers, inappropriate prescription practices and a significant increase in the number of immunocompromised individuals all contribute to a larger population who are highly susceptible to bacterial infection and to a frightening increase in both infection rates and drug resistance. This, together with the lack of new drugs being developed creates an urgent need for the appropriate molecular sciences research that underpins the difficult and longer-term studies required to develop antibiotics.
Using modern technologies in chemo-bioinformatics, proteomics, molecular and structural biology, computational chemistry and screening technologies we aim to identify ligands and inhibitors (hit and lead compounds) with the potential to drive the discovery of improved therapies for Gram-negative infections.
In St. Andrews we have solved the structures of more than 50 of AEROPATHs targets. Further studies on a selection of these targets has yielded structural information on numerous inhibitor-complexes, the first step in the development of a new antimicrobial.
RmlA
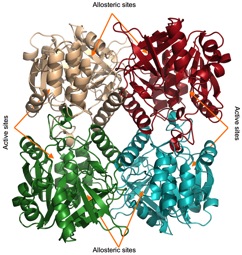
Rmla (glucose-1-phosphate thymidylyltransferase) is essential for the virulence of P. aeruginosa. A high-throughput compound screen was run in collaboration with the Drug Discovery Unit and the University of Dundee to identify potential inhibitors. The top hits from this screen allowed a rational drug design methodology to be followed to improve the potency of a series of compounds, using x-ray crystallography and synthetic chemistry. Interestingly, the compounds were found to bind at an allosteric site in the tetrameric RmlA, locking the quaternary structure in an inactive conformation and preventing the binding of substrate at the active site.
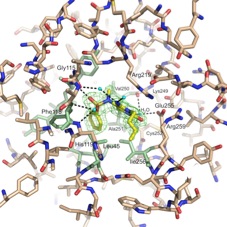
Fatty Acid Biosynthesis
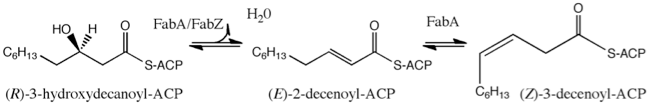
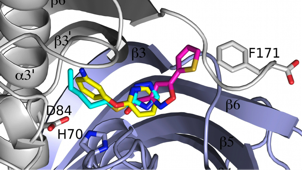
Fatty acid biosynthesis is an essential component of metabolism in both eukaryotes and prokaryotes. The fatty acid biosynthetic pathway of Gram-negative bacteria is an established therapeutic target. Two homologous enzymes FabA and FabZ catalyze a key step in fatty acid biosynthesis; both dehydrate hydroxyacyl fatty acids that are coupled via a phosphopantetheine to an acyl carrier protein (ACP). The resulting trans-2-enoyl ACP is further polymerized in a processive manner, FabA however carries out a second reaction involving isomerization of trans-2-enoyl fatty acid to cis-3-enoyl fatty acid. We have solved the structure of Pseudomonas aeruginosa FabA with a substrate allowing detailed molecular insight into the interactions of the active site. This has allowed a detailed examination of the factors governing the second catalytic step. We have also determined the structure of FabA in complex with small molecules (so called fragments). These small molecules bind at different points of the active site and form the basis for a rational inhibitor design program.